Appendix A: Optimizing Performance and Parallelization¶
Performance optimization on modern multi-core machines is a complex subject.
Know your Linear Algebra Library¶
The optical propagation calculations in POPPY are dominated by a small number
of matrix algebra calls, primarily np.dot with a side order of
np.outer. numpy relies heavily on ATLAS (A Tuned LaPACK and BLAS
library ) to perform its linear algebra
calculations, but for reasons of portability many distributed copies of numpy
do not have an ATLAS that has been compiled with all the CPU optimization
tricks turned on.
Whether or not numpy is linked against an optimized
linear algebra library can make a huge difference in execution speed, with
speedups of an order of magnitude or more. You definitely want to make sure
that your numpy is using such a library:
- Apple’s Accelerate framework (a.k.a. vecLib) provides a highly tuned copy of BLAS and LAPACK on any Mac, right out of the box.
- OpenBLAS is recommended on Linux.
- The Intel Math Kernel Library (MKL) Optimizations is available as an add-on from Continuum Analytics to their Anaconda Python distribution. This requires a commercial license for a small fee.
Numpy is statically linked at compile time against a given copy of BLAS. Switching backends generally requires recompiling numpy. (Note that if you use MacPorts on Mac OS to install numpy, it automatically uses Apple’s Accelerate framework for you. Nice!)
Various stackoverflow,
quora,
and twitter posts
suggest that OpenBLAS, MKL, and Accelerate all have very similar performance,
so as long as your numpy is using one of those three you should be in good
shape.
Parallelized Calculations¶
POPPY can parallelize calculations in two different ways:
- Using Python’s built-in
multiprocessingpackage to launch many additional Python processes, each of which calculates a different wavelength.- Using the FFTW library for optimized accellerated Fourier transform calculations. FFTW is capable of sharing load across multiple processes via multiple threads.
One might think that using both of these together would result in the fastest possible speeds. However, in testing it appears that FFTW does not work reliably with multiprocessing for some unknown reason; this instability manifests itself as occasional fatal crashes of the Python process. For this reason it is recommended that one use either multiprocessing or FFTW, but not both.
For most users, the recommended choice is using multiprocessing.
FFTW only helps for FFT-based calculations, such as some coronagraphic or spectroscopic calculations. Calculations that use only discrete matrix Fourier transforms are not helped by FFTW. Furthermore, baseline testing indicates that in many cases, just running multiple Python processes is in fact significantly faster than using FFTW, even for coronagraphic calculations using FFTs.
There is one slight tradeoff in using multiprocessing: When running in this mode, POPPY cannot display plots of the calculation’s work in progress, for instance the intermediate optical planes. (This is because the background Python processes can’t write to any Matplotlib display windows owned by the foreground process.) One can still retrieve the intermediate optical planes after the multiprocess calculation is complete and examine them then; you just can’t see plots displayed on screen as the calculation is proceeding. Of course, the calculation ought to proceed faster overall if you’re using multiple processes!
Warning
On Mac OS X, for Python < 2.7, multiprocessing is not compatible with Apple’s Accelerate framework mentioned above, due to the non-POSIX-compliant manner in which multiprocessing forks new processes. See https://github.com/spacetelescope/poppy/issues/23 and https://github.com/numpy/numpy/issues/5752 for discussion. Python 3.4 provides an improved method of starting new processes that removes this limitation.
If you want to use multiprocessing with Apple’s Accelerate framework, you must upgrade to Python 3.4+. POPPY will raise an exception if you try to start a multiprocess calculation and numpy is linked to Accelerate on earlier versions of Python.
This is likely related to the intermittent crashes some users have reported with multiprocessing and FFTW; that combination may also prove more stable on Python 3.4 but this has not been extensively tested yet.
The configuration options to enable multiprocessing live under poppy.conf, and use the Astropy configuration framework. Enable them as follows:
>>> import poppy
>>> poppy.conf.use_multiprocessing = True
>>> poppy.conf.use_fftw = False
One caveat with running multiple processes is that the memory demands can become substantial for large oversampling factors. For instance, a 1024-pixel-across pupil with oversampling=4 results in arrays that are 256 MB each. Several such arrays are needed in memory per calculation, with peak memory utilization reaching ~ 1 GB per process for oversampling=4 and over 4 GB per process for oversamping=8.
Thus, if running on a 16-core computer, ensure at least 32 GB of RAM are available before using one process per core. If you are constrained by the amount of RAM available, you may experience better performance using fewer processes than the number of processor cores in your computer.
By default, POPPY attempts to automatically choose a number of processes based on available CPUs and free memory that will optimize computation speed without exhausting available RAM. This is implemented in the function poppy.utils.estimate_optimal_nprocesses().
If desired, the number of processes can be explicitly specified:
>>> poppy.conf.n_processes = 5
Set this to zero to enable automatic selection via the estimate_optimal_nprocesses() function.
Comparison of Different Parallelization Methods¶
The following figure shows the comparison of single-process, single-process with FFTW, and multi-process calculations on a relatively high end 16-core Mac Pro. The calculations were done with WebbPSF, a PSF simulator for JWST that uses POPPY to perform computations.
The horizontal axis shows increasing detail of calculation via higher oversampling, while the vertical axis shows computation time. Note the very different Y-axis scales for the two figures; coronagraphic calculations take much longer than direct imaging!
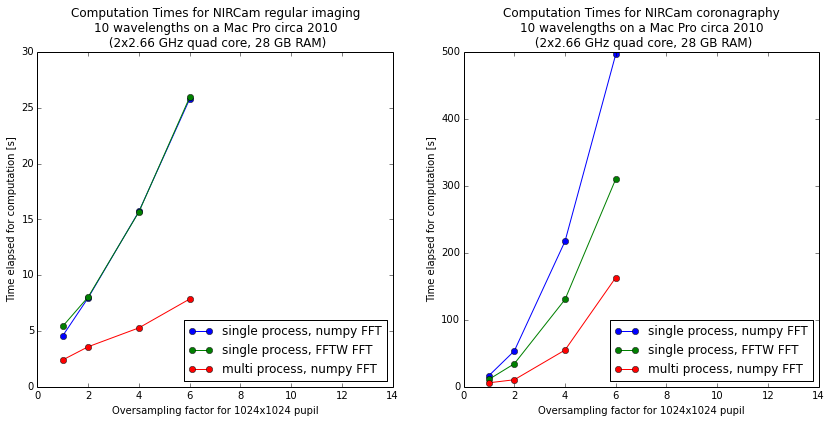
Using multiple Python processes is the clear winner for most workloads. Explore the options to find what works best for your particular calculations and computer setup.